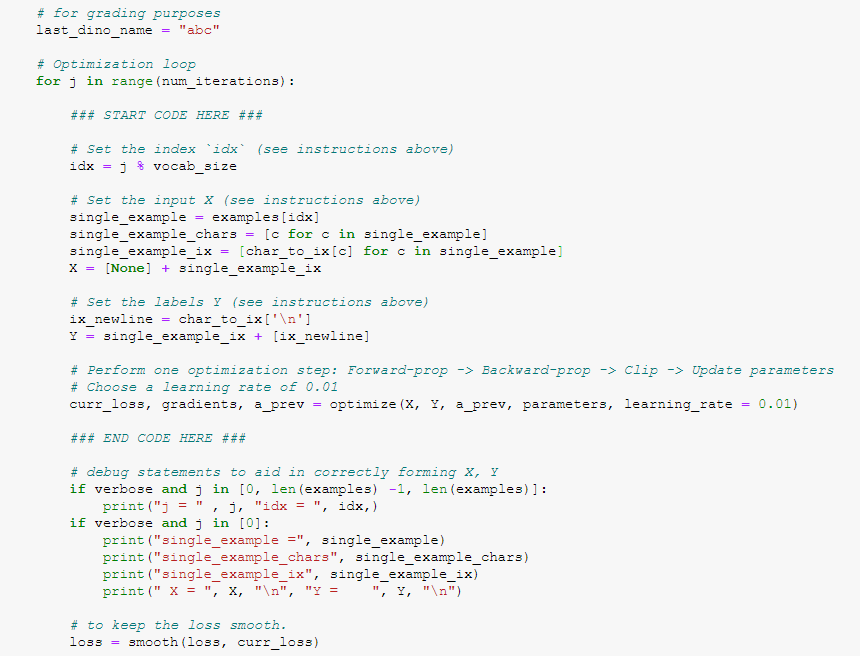
j = 0 idx = 0
single_example = turiasaurus
single_example_chars ['t', 'u', 'r', 'i', 'a', 's', 'a', 'u', 'r', 'u', 's']
single_example_ix [20, 21, 18, 9, 1, 19, 1, 21, 18, 21, 19]
X = [None, [20, 21, 18, 9, 1, 19, 1, 21, 18, 21, 19]]
Y = [[20, 21, 18, 9, 1, 19, 1, 21, 18, 21, 19], 0]
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
<ipython-input-16-725c093d6b91> in <module>
----> 1 parameters, last_name = model(data.split("\n"), ix_to_char, char_to_ix, 22001, verbose = True)
2
3 assert last_name == 'Trodonosaurus\n', "Wrong expected output"
4 print("\033[92mAll tests passed!")
<ipython-input-15-ad9b5cfee8e5> in model(data_x, ix_to_char, char_to_ix, num_iterations, n_a, dino_names, vocab_size, verbose)
82 if j % 2000 == 0:
83
---> 84 print('Iteration: %d, Loss: %f' % (j, loss) + '\n')
85
86 # The number of dinosaur names to print
TypeError: only size-1 arrays can be converted to Python scalars
Sir this is the error I am facing